Features
Colony based
Compatible with many microbial tests that require pure culture or viable cells
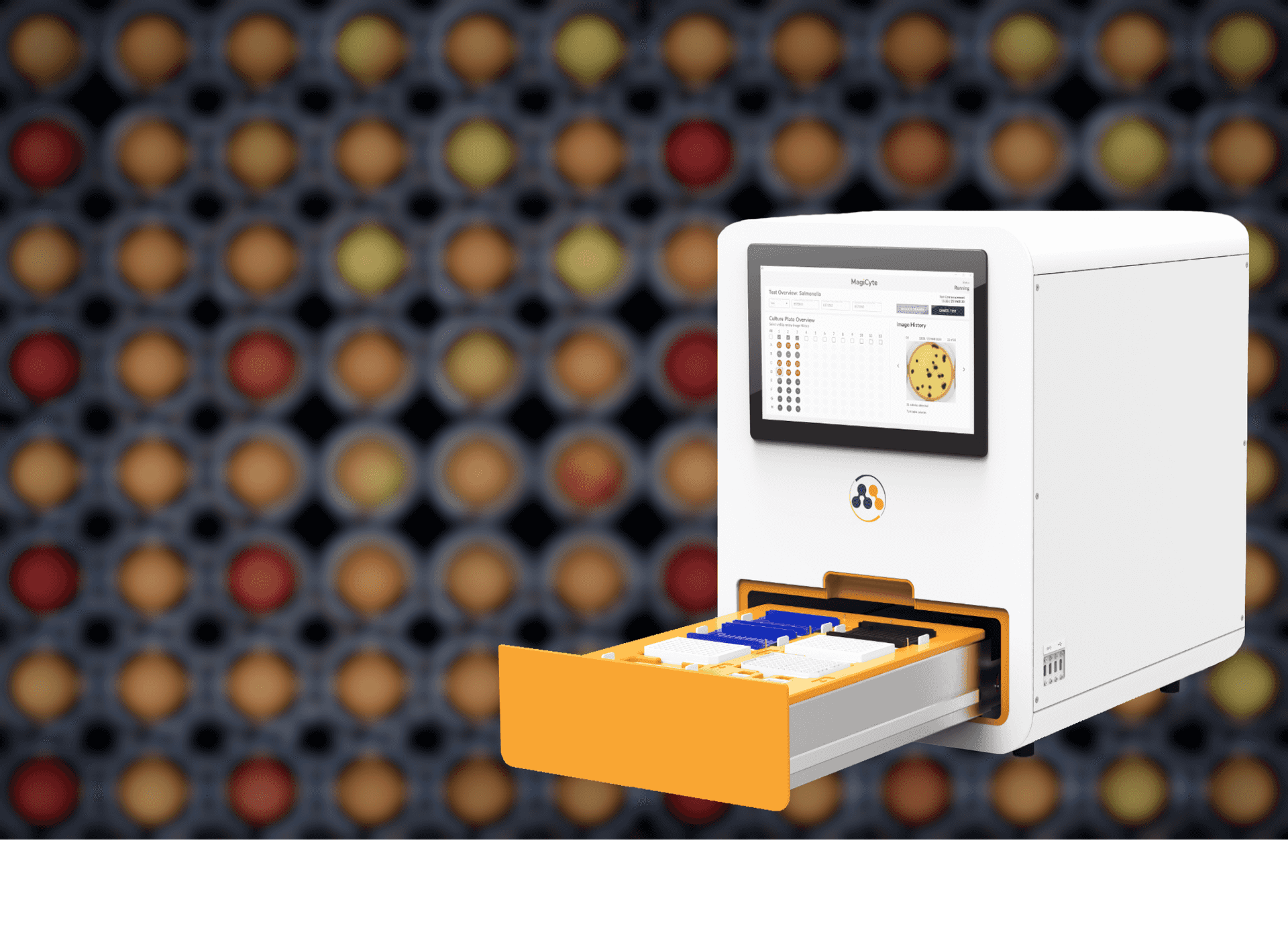
Full automation
A true walkaway solution: samples in, results and colonies out.
High throughput
Up to 96 samples per run
Easy workflow
Easy to use and set up
Rapid test
Results and colonies can be available in 24 hours, allowing downstream tests to start days earlier.
Accurate & reliable
Reduce human errors and contamination risks
Small footprint
Benchtop instrument and 96-well microplates. No more Petri dishes!
Lower cost
Save your lab as much as 50% per test
Why Microbial Culture?
Culture Methods
Take Salmonella testing in food as an example. Culture methods, especially reference methods such as FDA BAM, USDA MLG and ISO, are widely used for Salmonella detection and isolation. Unlike detection-only molecular methods, culture methods are indispensable for obtaining discrete Salmonella colonies, which are needed by numerous downstream confirmation and characterization technologies.
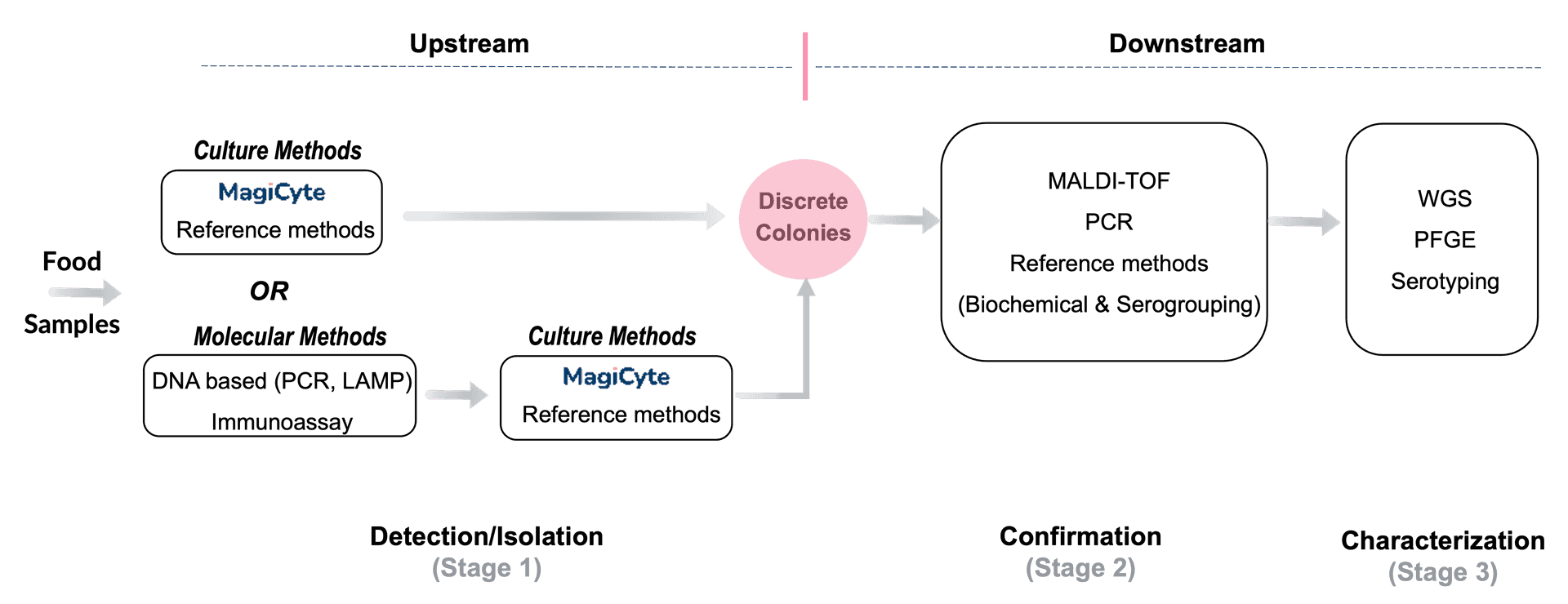
MagiCyte vs. Other Upstream Methods
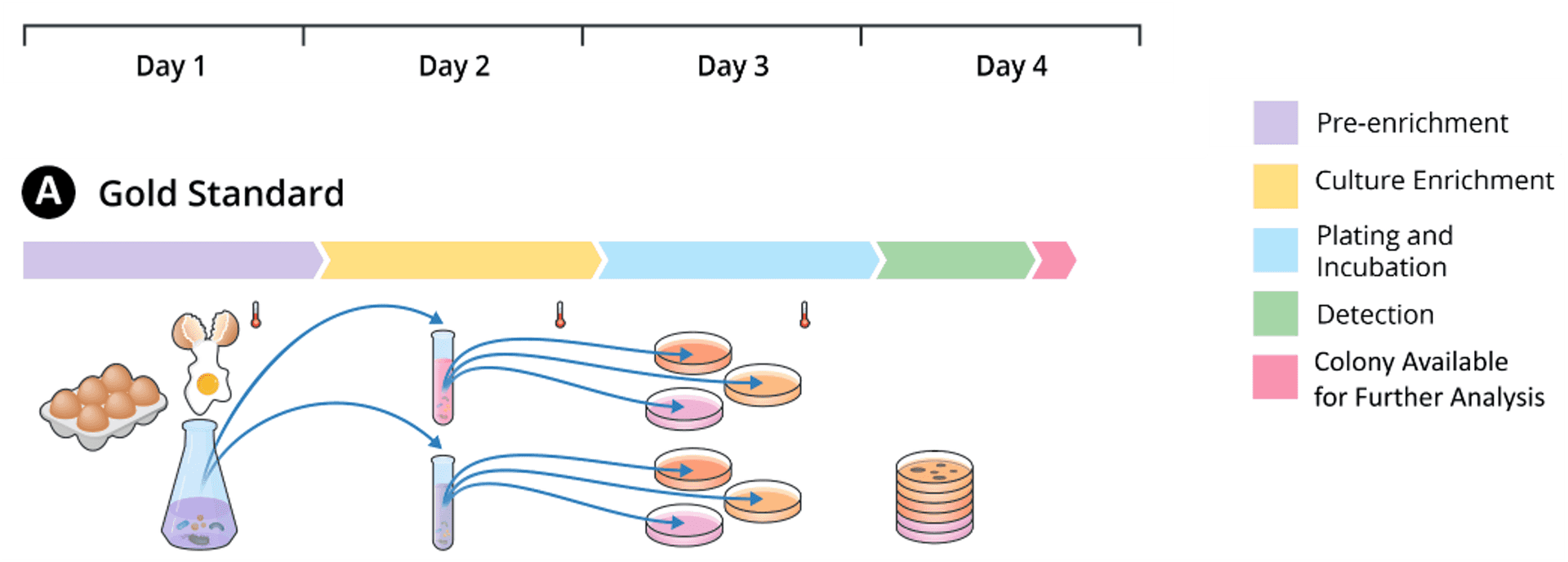
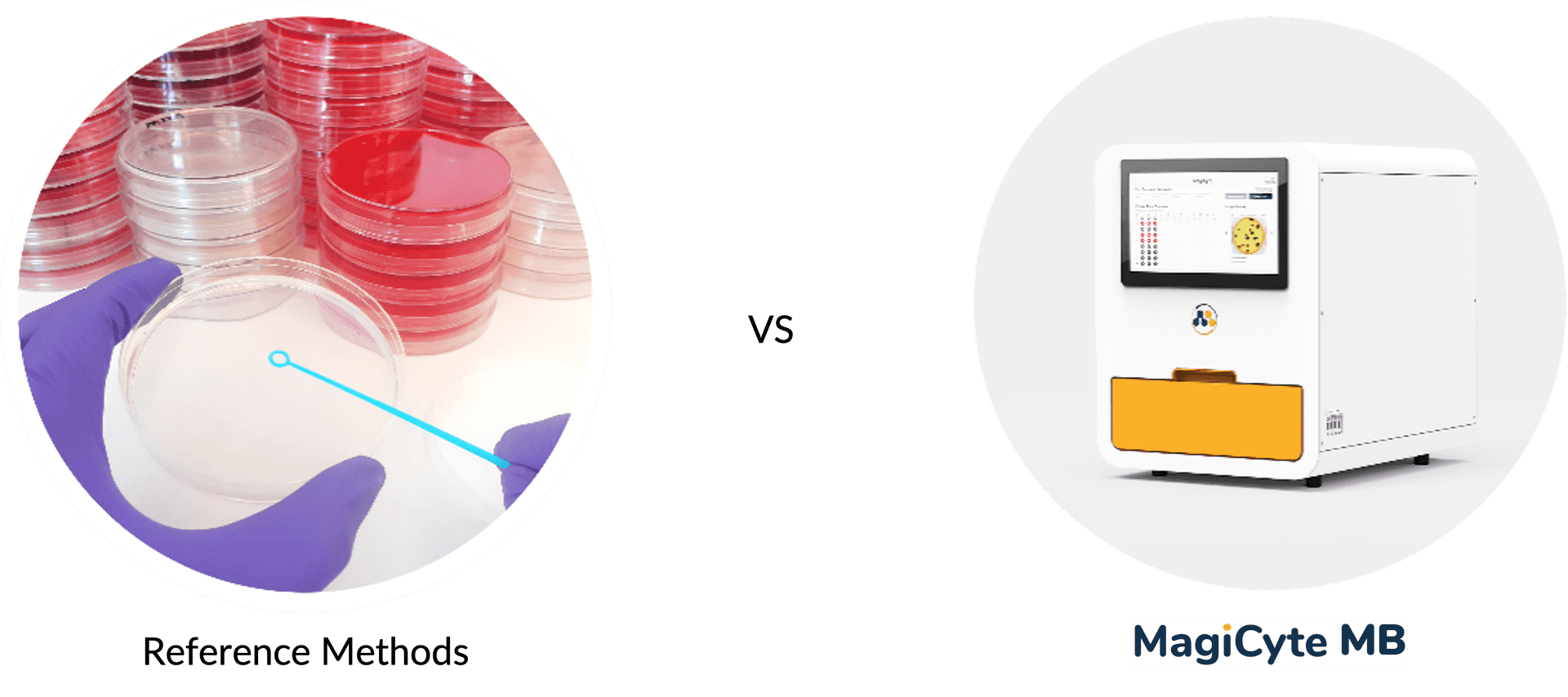
Reference Methods
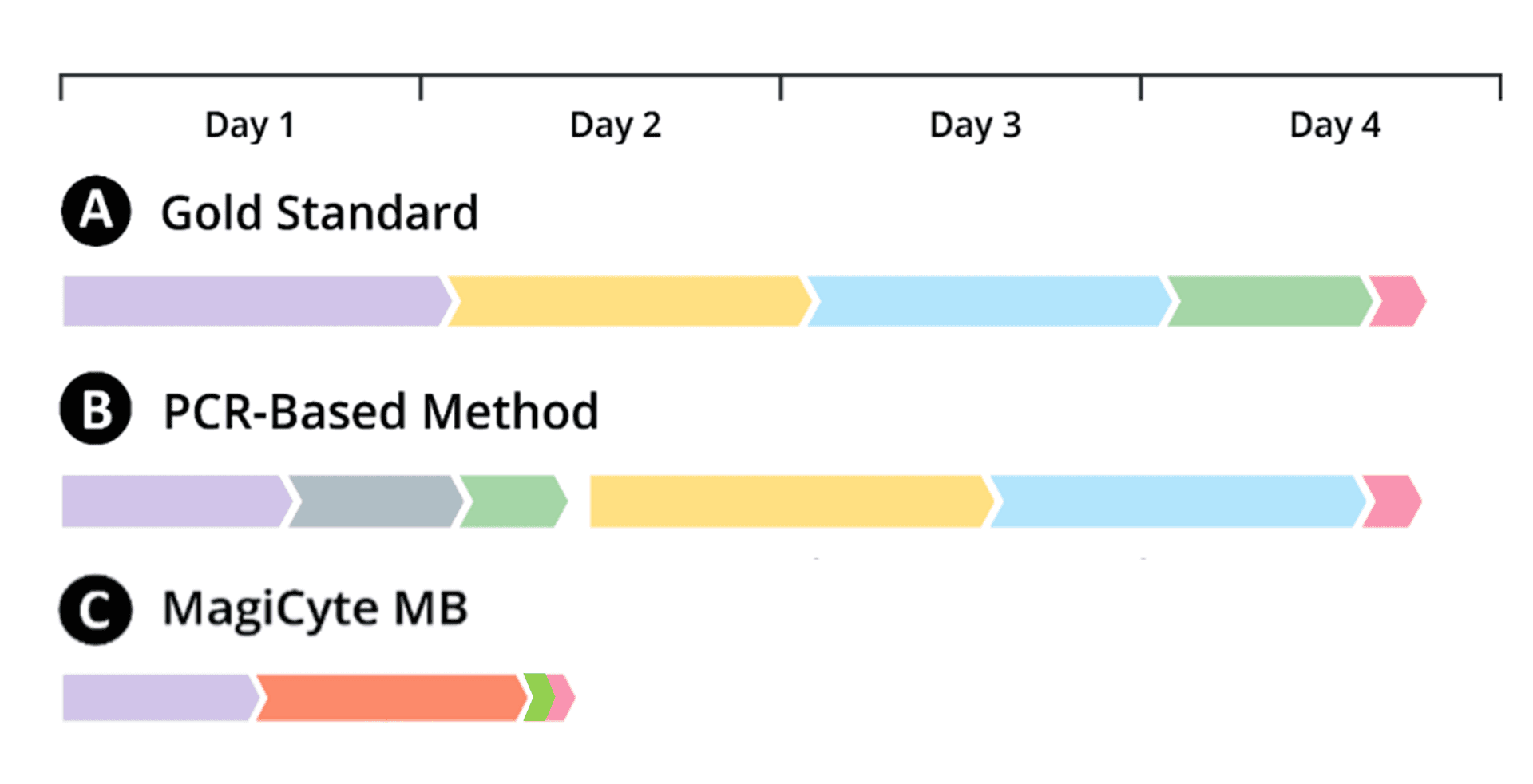
Reference methods for Salmonella detection and isolation usually include multiple time-consuming and resource-intensive culture steps. For example, FDA BAM requires three culture steps with each step taking about one day: first in non-selective enrichment media, then in selective enrichment broths, and finally in selective agars. As a result, labs have to wait until Day 4 to get detection results and obtain colonies before they can start downstream analyses.
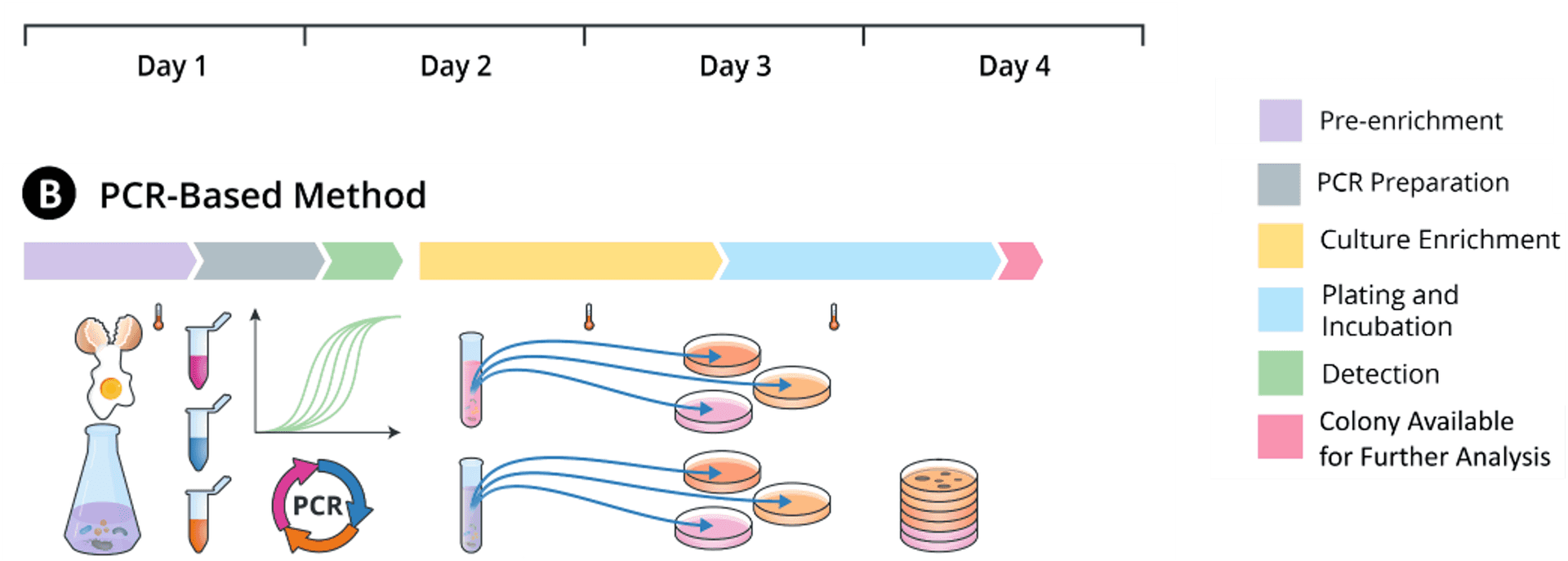
PCR
With molecular methods such as PCR, labs can get detection results faster. But for presumptive positive samples, labs need isolates for downstream analyses and still have to wait until Day 4 due to the subsequent use of the FDA BAM method.
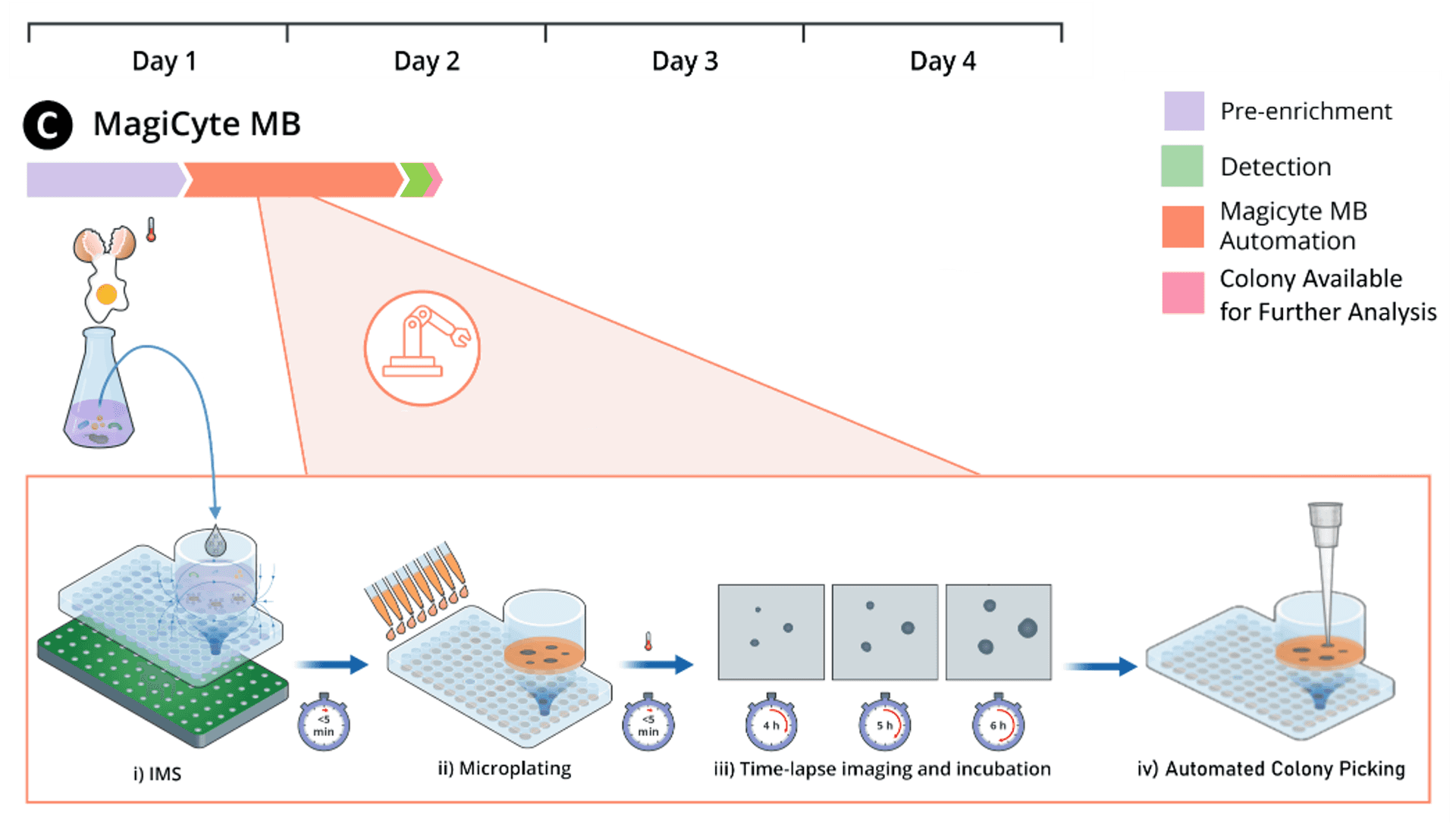
MagiCyte MB
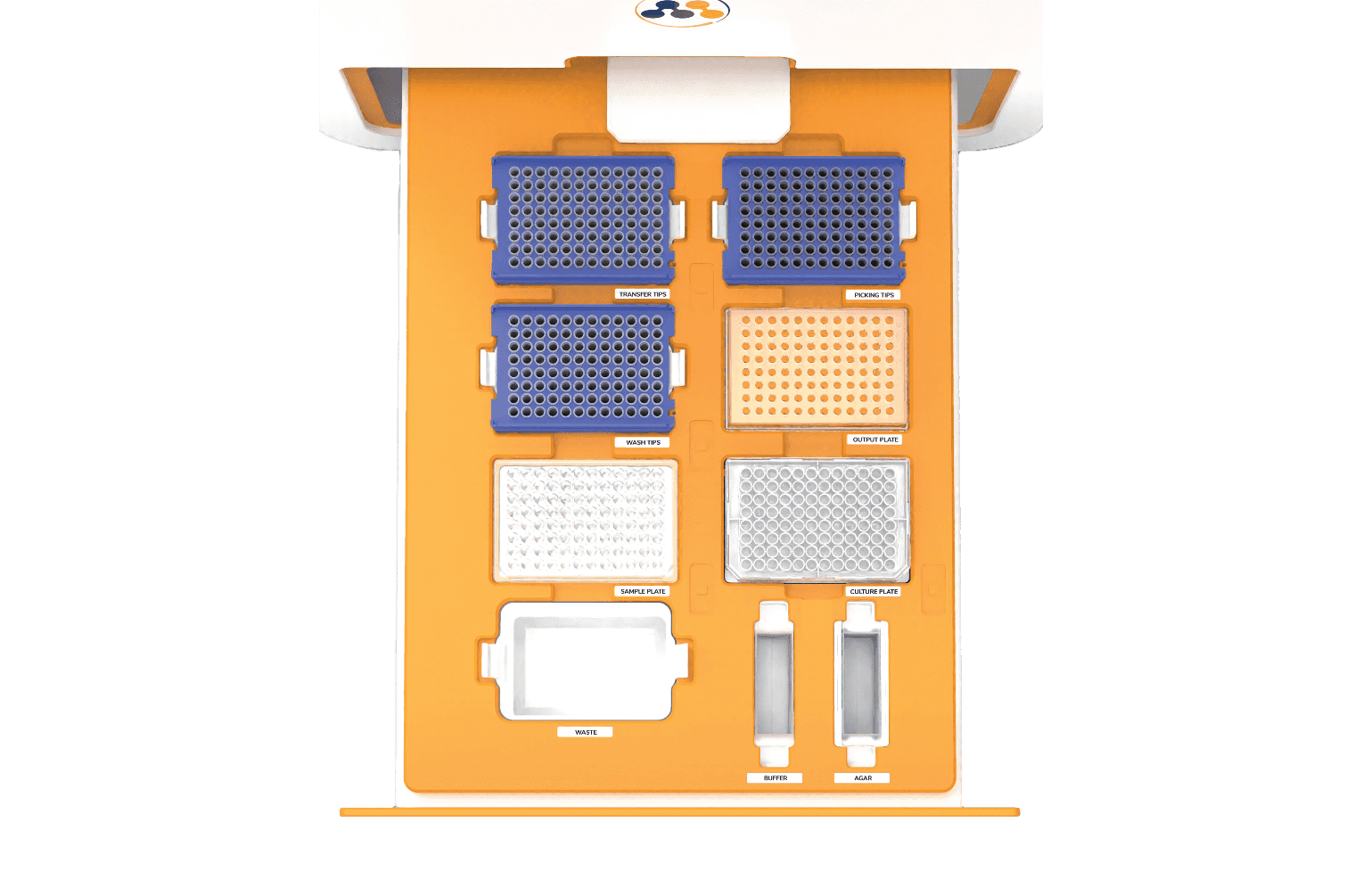
In MagiCyte MB, Salmonella are first separated from the samples (which might contain competing microbiota, chemical, inhibitors, etc.) by magnetic beads coated with anti-Salmonella antibodies through a selective enrichment step called immunomagnetic separation (IMS). The separated Salmonella are then plated into selective agar where the microcolonies' morphology are analyzed in real time for rapid and reliable detection and enumeration. Top-ranked discrete colonies are automatically picked and placed in an "output" plate for downstream analyses.
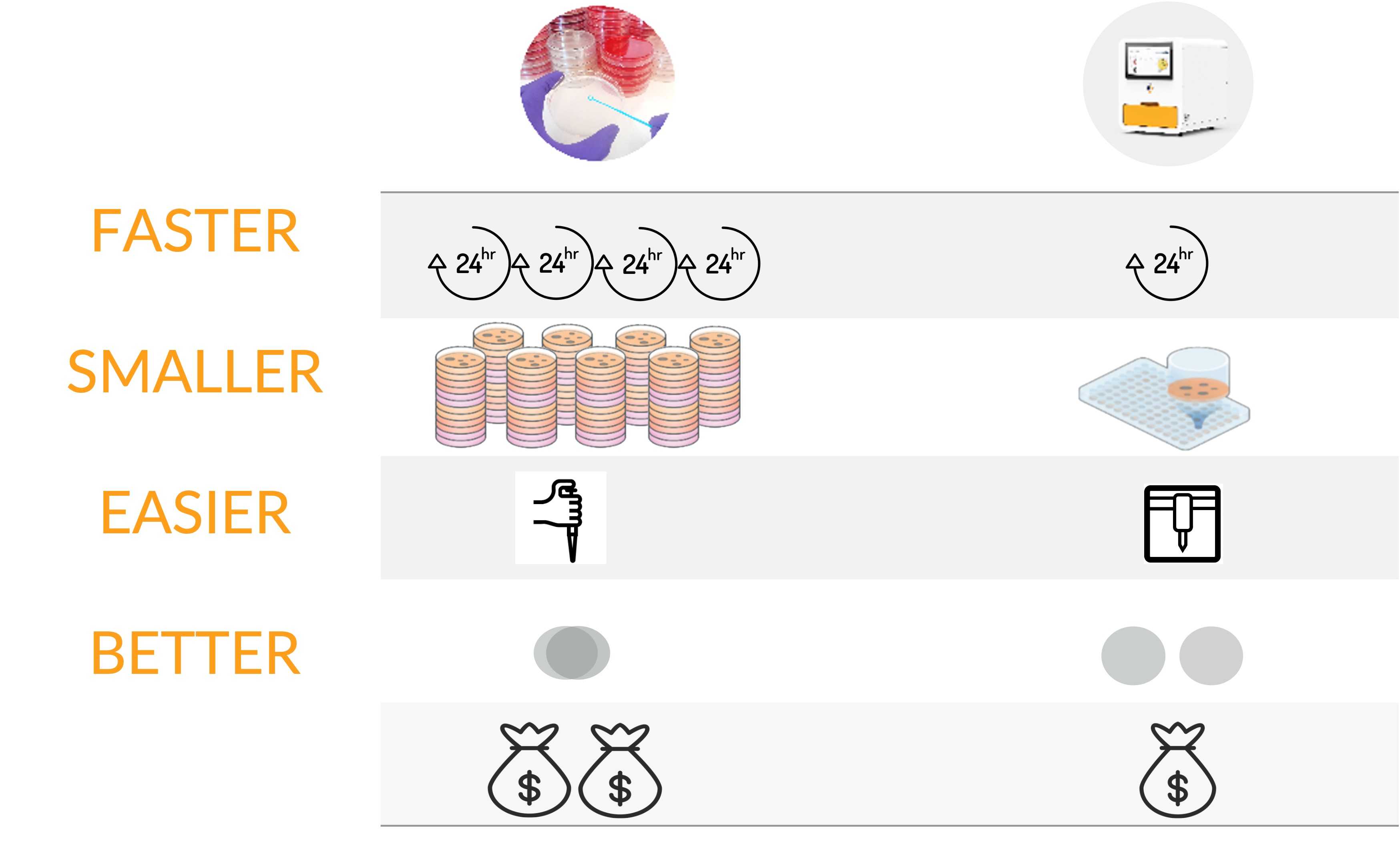
FASTER. SMALLER. EASIER. BETTER.
FASTER
Salmonella doubling time is less than 30 minutes when incubated at 37 °C, i.e., in a few hours, their colonies are easily visible under a low-resolution microscope and contain enough cells for downstream analyses. Why wait 4 days? MagiCyte obtains colonies days earlier and allows downstream tests to start much sooner.
SMALLER
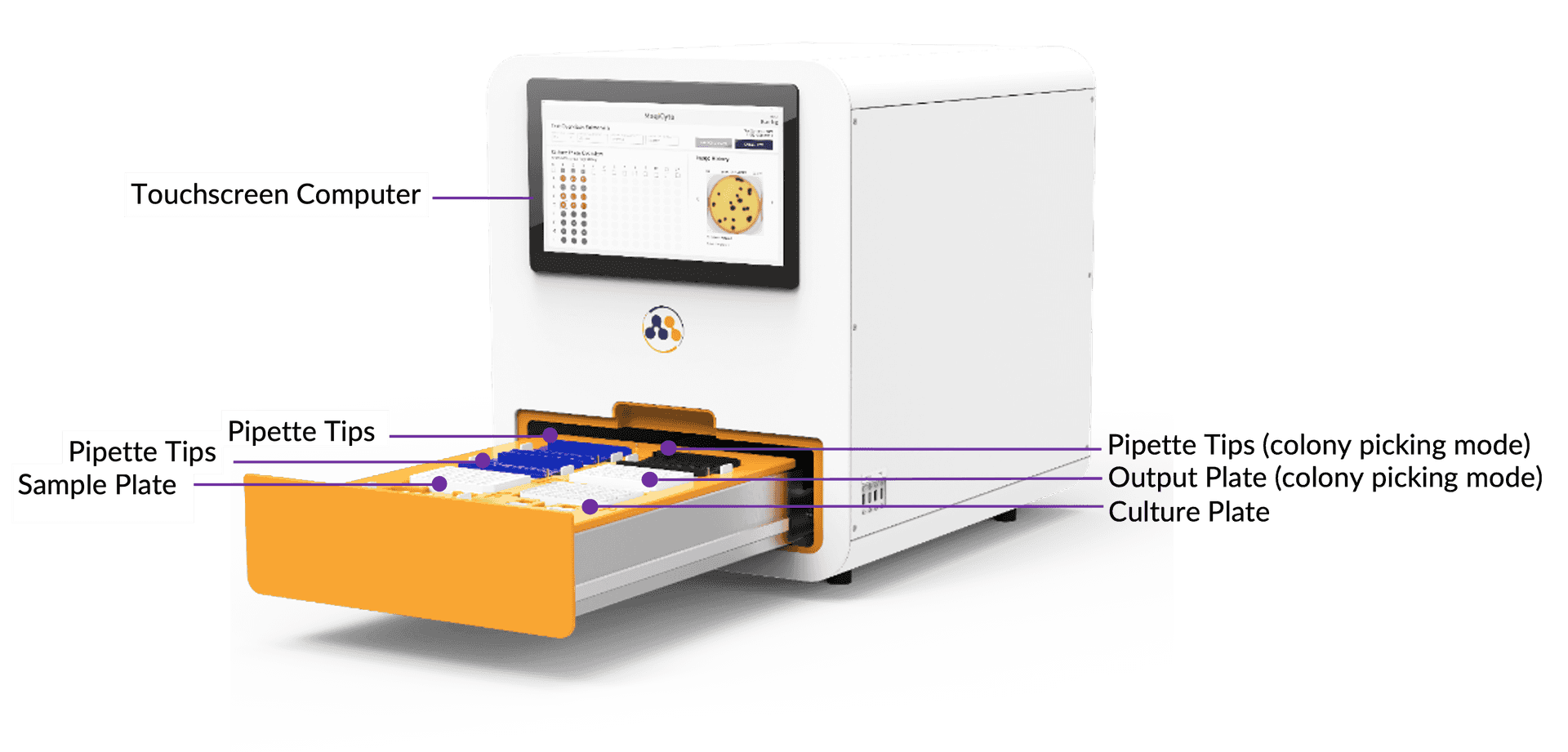
Why use Petri dishes for microbial culture? By replacing Petri dishes with a standard 96-well plate, MagiCyte MB significantly saves space and materials.
EASIER
A true walk away solution: samples in, results & colonies out.

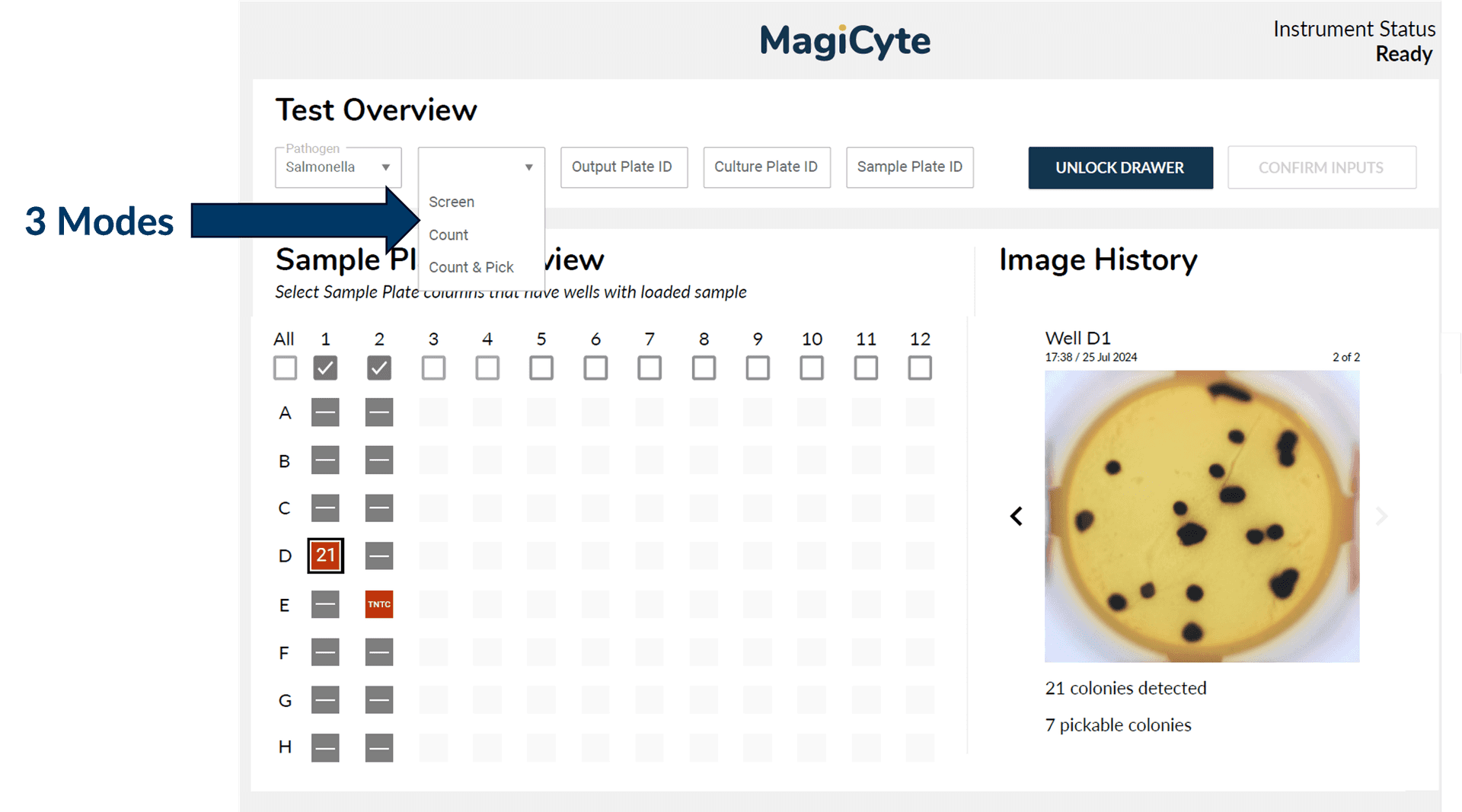
Screening? Enumeration? Isolation? Just select the mode on the intuitive software user interface and MagiCyte MB will do it for you.
BETTER
How are the colonies formed? Are the colonies discrete? Real time colony analysis by AI provides the answers to achieve accurate detection, enumeration and colony picking. Automation and standardization make the process reliable and repeatable.
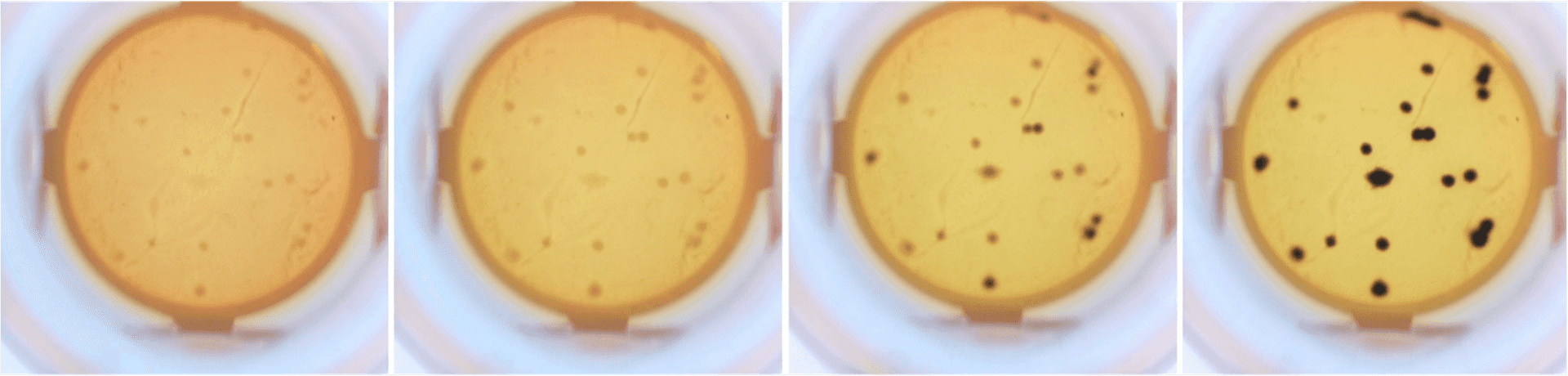
How does MagiCyte MB stack up?